Research
Our research focuses on engineering microbial cell-based systems to perform complex tasks and to produce renewable fuels, chemicals, and advanced materials.
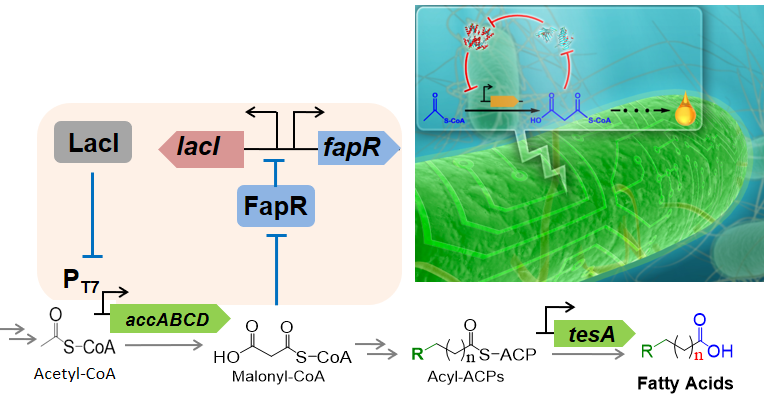
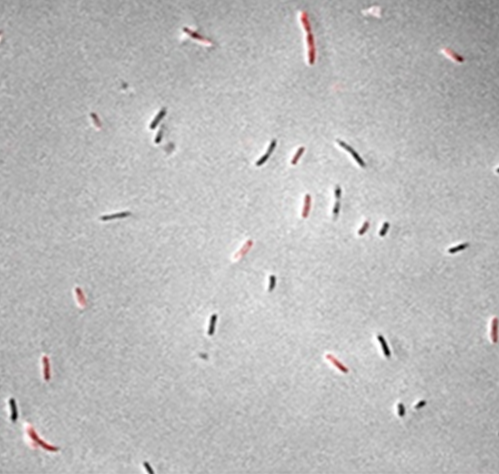
1. Metabolic Dynamics and Heterogeneity : (synthetic biology, metabolic engineering) Nature uses a suite of delicate regulatory systems to control both gene expression and pathway flux in response to environmental signals. When engineering microbial cells to produce fuels, chemicals, and materials, it is important to rewire the natural regulatory network, which was evolved for cell growth and replication, but not to over-produce any target compound. We are interested in developing tools to dissect the complex regulatory network of natural systems and to design synthetic regulatory systems that allow cell to control engineered pathways dynamically (Fig left). In addition, non-genetic, cell-to-cell variations in protein and metabolite concentrations cause large heterogeneity in single cell biosynthetic performance and dramatically affect ensemble product yields and titers (Fig right). We are interested in understanding the origin of metabolic noise and developing tools to regulate metabolic variation.
Representative papers: Liu D, Zhang F., Metabolic Feedback Circuits Provide Rapid Control of Metabolite Dynamics. ACS Syn Biol. 7 (2), 347–356, doi: 10.1021/acssynbio.7b00342 (2018). Schmitz AC, Hartline CJ, Zhang F, Engineering Microbial Metabolite Dynamics and Hetergeneity. Biotech J . (2017) Sep 13. doi: 10.1002/biot.201700422. Xiao Y., Bowen C., Liu D., Zhang F., Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat Chem Biol. 12, 339, (2016). Liu, D., Xiao, Y., Evans, B., Zhang, F., Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator. ACS Synth Biol. 4, 132, (2015). |
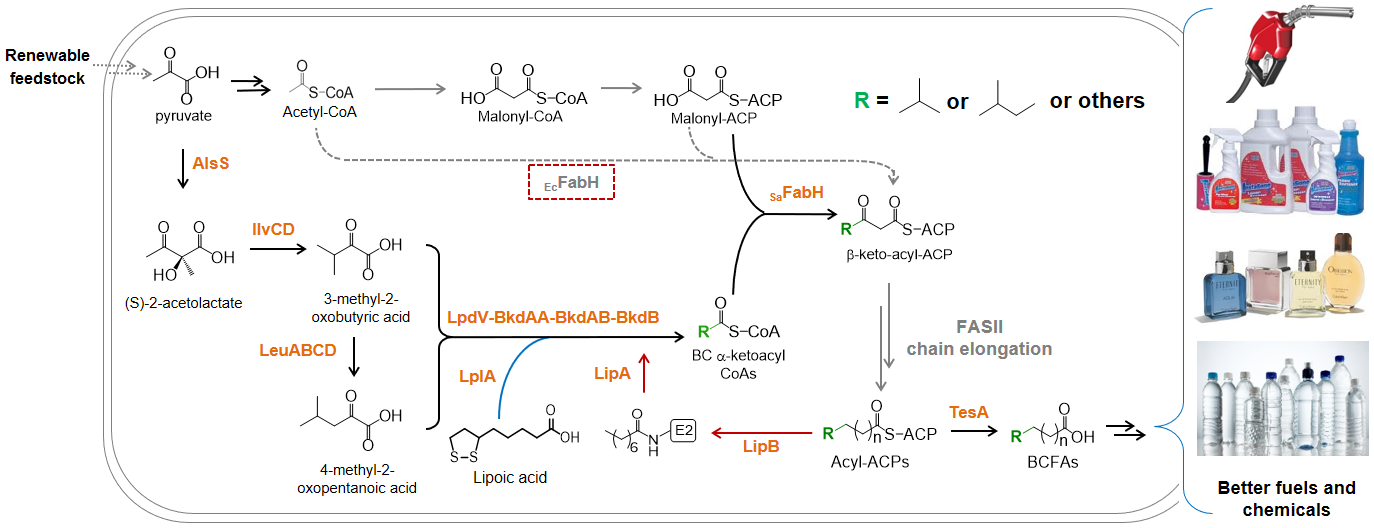
2. Advanced Biofuels (metabolic engineering & synthetic biology) To meet the increasing demands of sustainable transportation energy, we aim to engineer microbes to produce advanced biofuels that could be readily used in current engines. One of the biggest challenges is to biosynthesize non-natural molecules that have structures exactly the same or highly similar to the fuels currently derived from petroleum. We are interested in constructing novel metabolic pathways to produce these compounds. In addition, we are also developing synthetic biology tools to improve the titer, yield, and productivity of the advanced biofuels.
Representative papers: Jiang W., Qiao J.B., Bentley G.J., Liu D., Zhang F., Modular pathway engineering for the Production of Branched-Chain Fatty Alcohols. Biotechnol Biofuels. 10:244. doi: 10.1186/s13068-017-0936-4. eCollection (2017). Bentley G.J., Jiang W., Guamán L.P., Xiao Y., Zhang F., Engineering Escherichia coli to produce branched-chain fatty acids in high percentages. Metab Eng. 38, 148, (2016). Bowen C., Bonin J., Kogler A., Barba-Ostria C., Zhang F., Engineering Escherichia coli for conversion of glucose to medium-chain ω-hydroxy fatty acids and α,ω-dicarboxylic acids. ACS Syn Biol. 5, 200, (2016). Jiang W., Jiang Y., Bentley G.J., Liu D., Xiao Y., Zhang F., Enhanced production of branched-chain fatty acids by replacing β-ketoacyl-(acyl-carrier-protein) synthase III (FabH). Biotechnol Bioeng. 112, 1613, (2015). |
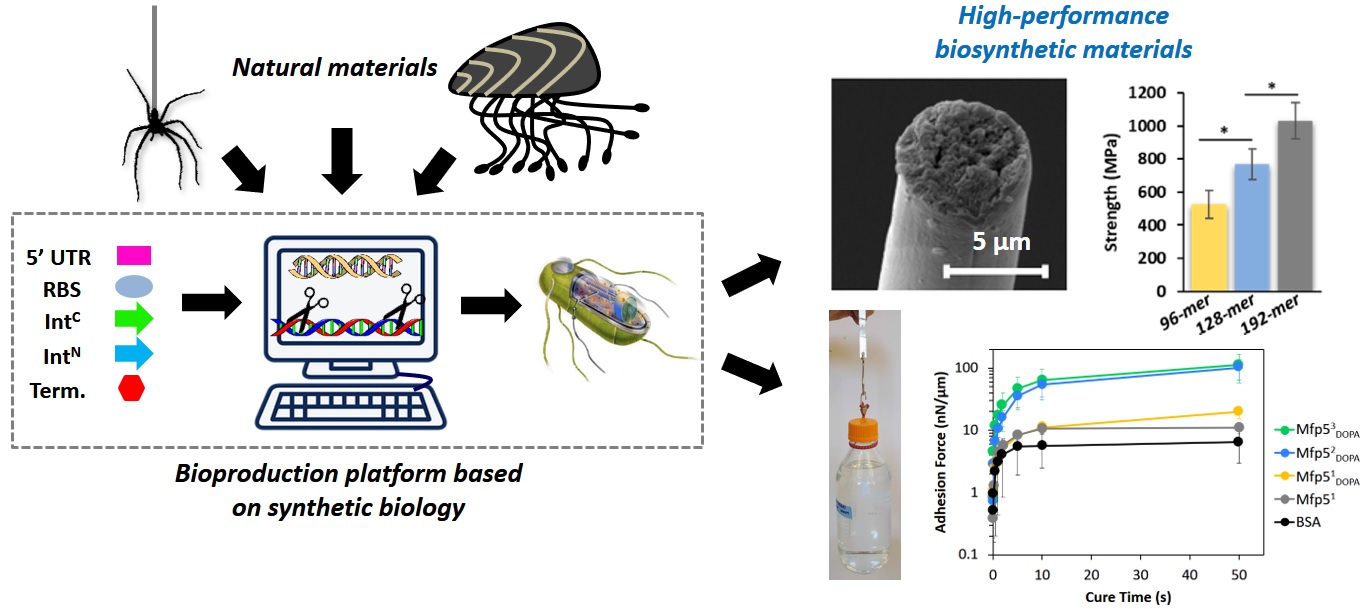
3. Advanced Materials (synthetic biology)
Representative papers: Bai W., Sargent C.J., Choi J., Pappu R.V., Zhang F.*, Covalently-assembled, single-chain protein nanostructures with ultra-high stability. Nature Communications, 10, 3317. doi://doi.org/10.1038/s41467-019-11285-8 (2019) Dai B., Sargent C.J., Gui X., Liu C., Zhang F., Fibril self-assembly of amyloid–spider silk block polypeptides. Biomacromolecules, 20(5):2015-2023. doi: 10.1021/acs.biomac.9b00218. (2019) Kim E., Dai B., Qiao J.B., Li W., Fortner J.D., Zhang F., Microbially Synthesized Repeats of Mussel Foot Protein Display Enhanced Underwater Adhesion. ACS Appl. Mater. Interfaces, 10 (49), 43003–43012 (2018) DOI: 10.1021/acsami.8b14890. Bowen C.H.*, Dai B.*, Sargent C.J., Bai W, Ladiwala P, Feng H., Huang W, Kaplan D.L., Galazka J.M., Zhang F., Recombinant Spidroins Replicate Mechanical Properties of Natural Spider Silk. Biomacromolecules, 19 (9), 3853–3860 (2018) DOI: 10.1021/acs.biomac.8b00980. |